Testing the equality of mean densities of temperature in Vietnam
DANOVA.qmd
library(dda)
library(fda.usc)
library(fdANOVA)
library(MVN)
library(tidyverse)
library(boot)
library(sf)
set.seed(10983)Preprocessing into density data
data(vietnam_temperature_dd)
data(vietnam_regions)
data(vietnam_provinces)
to_trend <- function(ddobj, t = seq_len(ncol(ddobj$coefs))) {
ddobj$coefs <- lm(t(ddobj$coefs) ~ t)$coefficients["t", ]
dd(clr = fd(ddobj$coefs, ddobj$basis))
}
vnt <- vietnam_temperature_dd |>
group_by(region, province) |>
summarise(t_max = list(c(t_max)), .groups = "drop") |>
mutate(t_max = lapply(t_max, to_trend)) |>
left_join(vietnam_regions, by = c("region" = "code")) |>
rename(region_name = name) |>
arrange(region, province)
class(vnt$t_max) <- c("ddl", "fdl", "list")
vnt |> plot_funs(t_max, color = region_name) +
facet_grid(vars(str_wrap(region_name, 14)), scales = "free_y") +
theme(legend.position = "none", axis.title.y = element_text(angle = -90)) +
geom_hline(yintercept = 1 / diff(vnt$t_max[[1]]$basis$rangeval), linetype = "dotted") +
labs(
x = "Temperature (deg. Celsius)",
y = "Density",
color = "Region"
)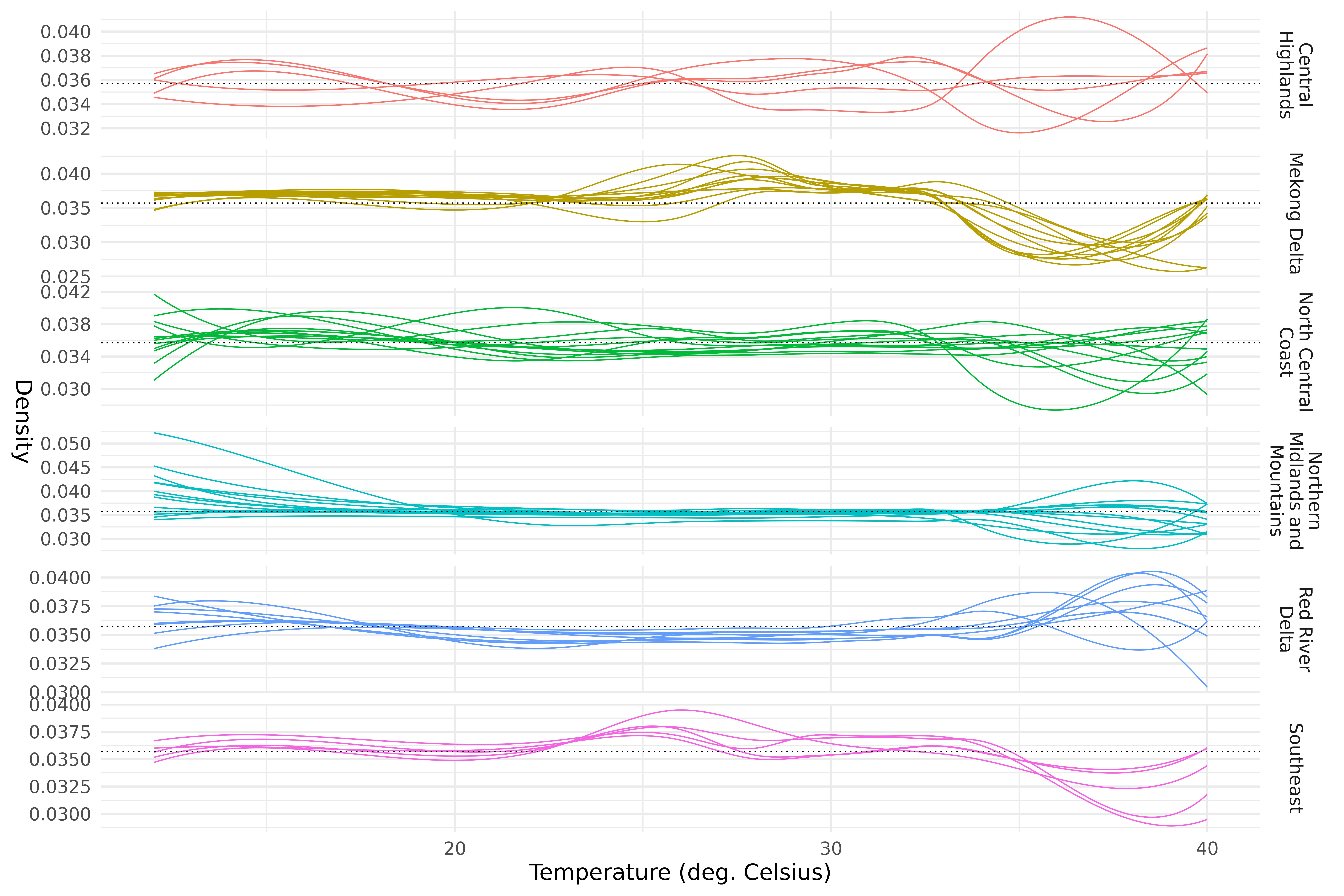
Density of maximum temperature per province.
vnt |> plot_funs(t_max, color = region_name) +
theme_legend_inside +
labs(
x = "Temperature (deg. Celsius)",
y = "Density",
color = "Region"
)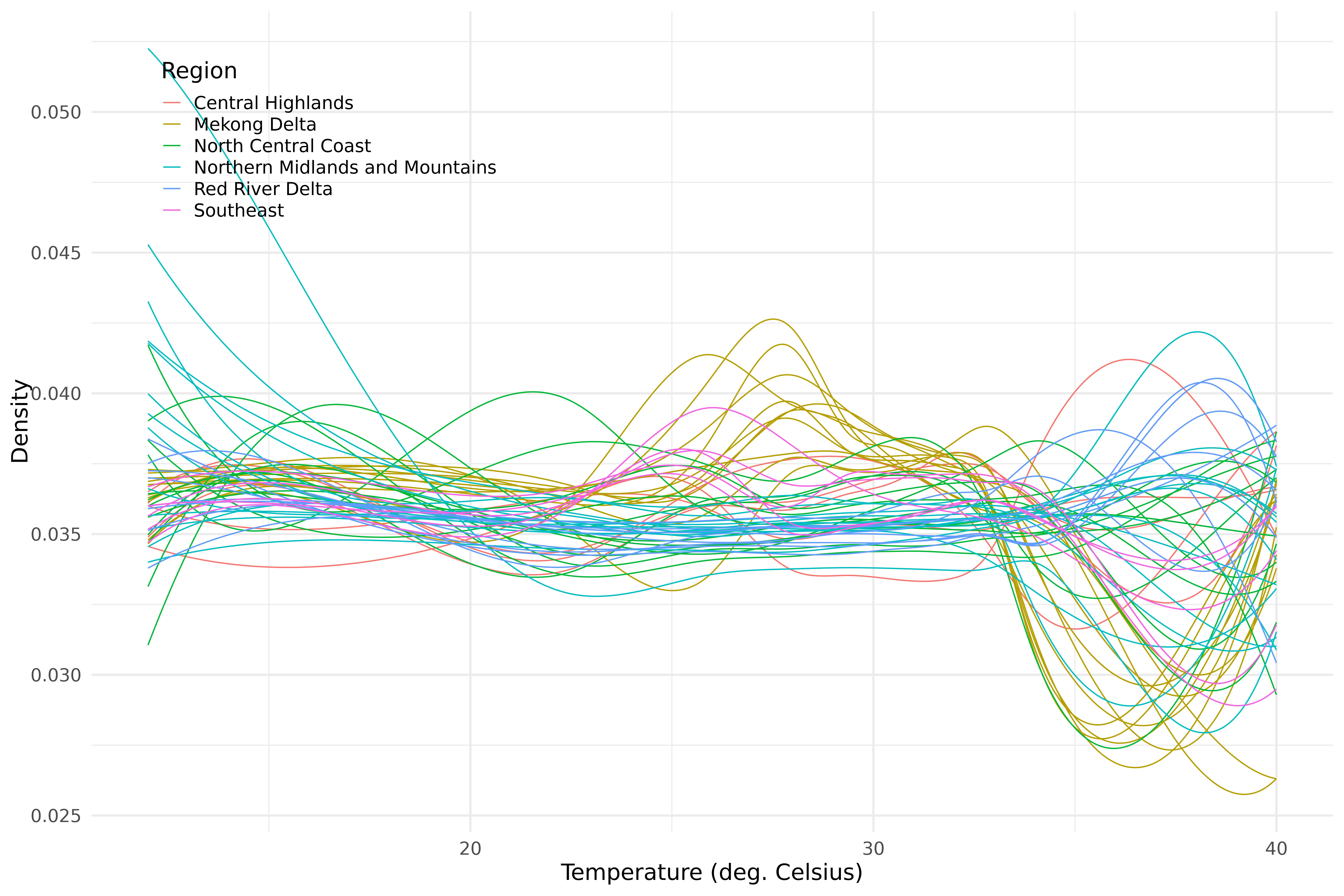
Density of maximum temperature per province.
vnt |>
pull(t_max) |>
mean() |>
plot() +
geom_hline(yintercept = 1 / diff(vnt$t_max[[1]]$basis$rangeval), color = "red", linetype = "dotted") +
labs(
x = "Temperature (deg. Celsius)",
y = "Density"
)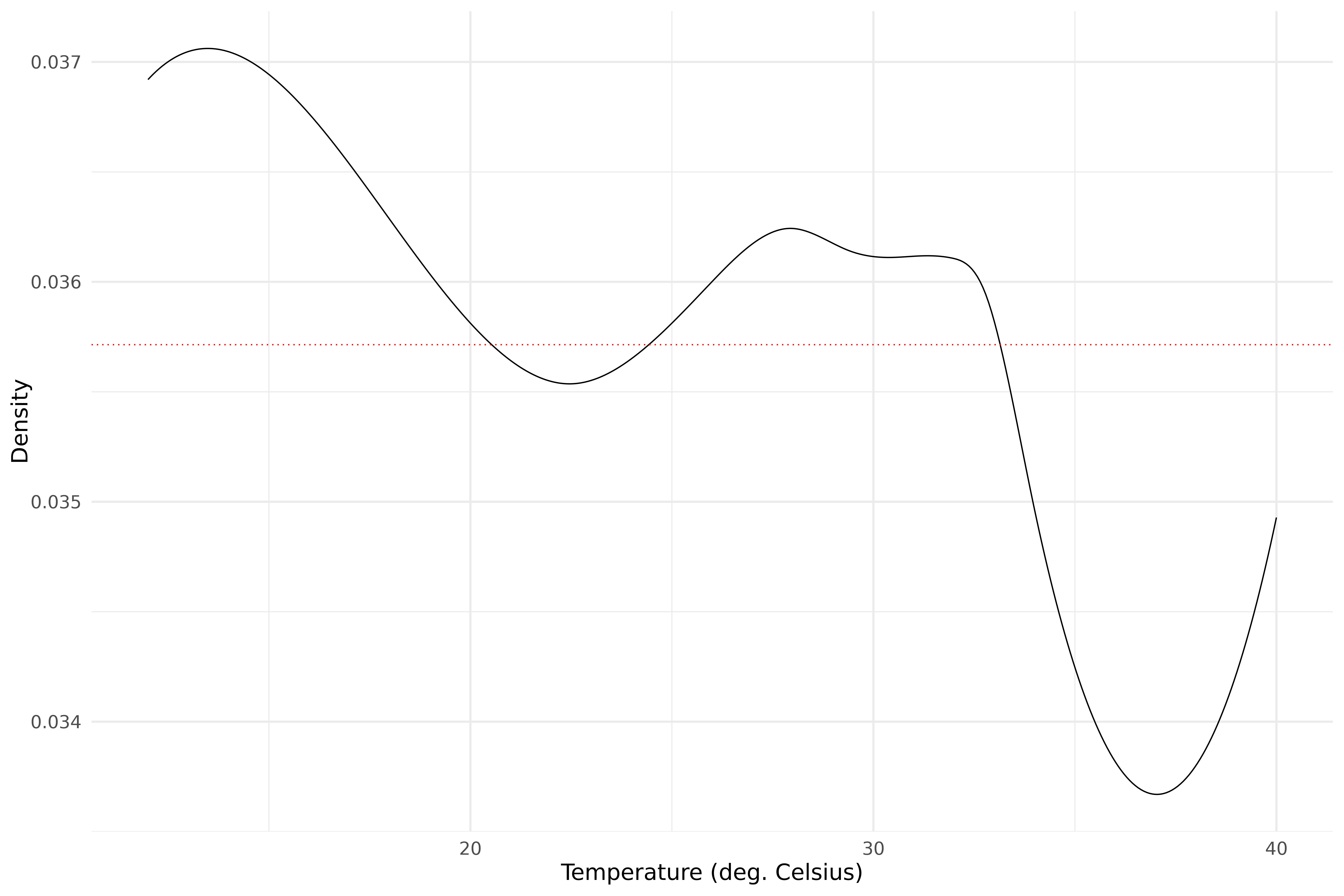
Mean annual trend of the maximum temperature distributions between 1987 and 2016.
vnt |>
pull(t_max) |>
mean() |>
as.fd() |>
plot() +
labs(
x = "Temperature (deg. Celsius)",
y = "clr"
)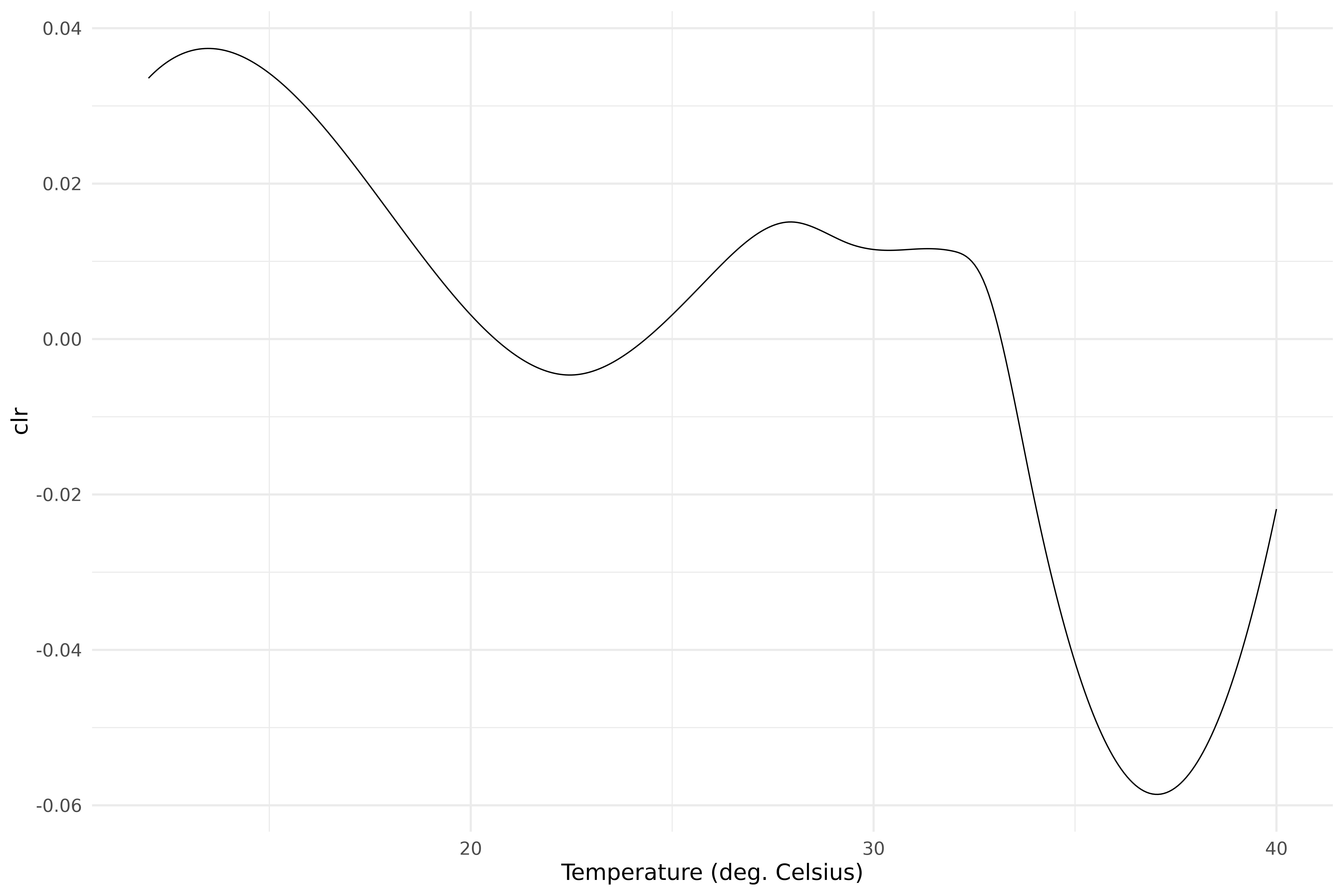
Centered log-ratio transform of the mean annual trend of maximum temperature distributions between 1987 and 2016.
Exploratory analysis by region
vietnam_provinces |>
left_join(vietnam_regions, by = c("region" = "code")) |>
rename(region_name = name) |>
ggplot(aes(fill = region_name)) +
geom_sf() +
scale_x_continuous(breaks = seq(from = -180, to = 180, by = 2)) +
labs(x = "Longitude", y = "Latitude", fill = "Region")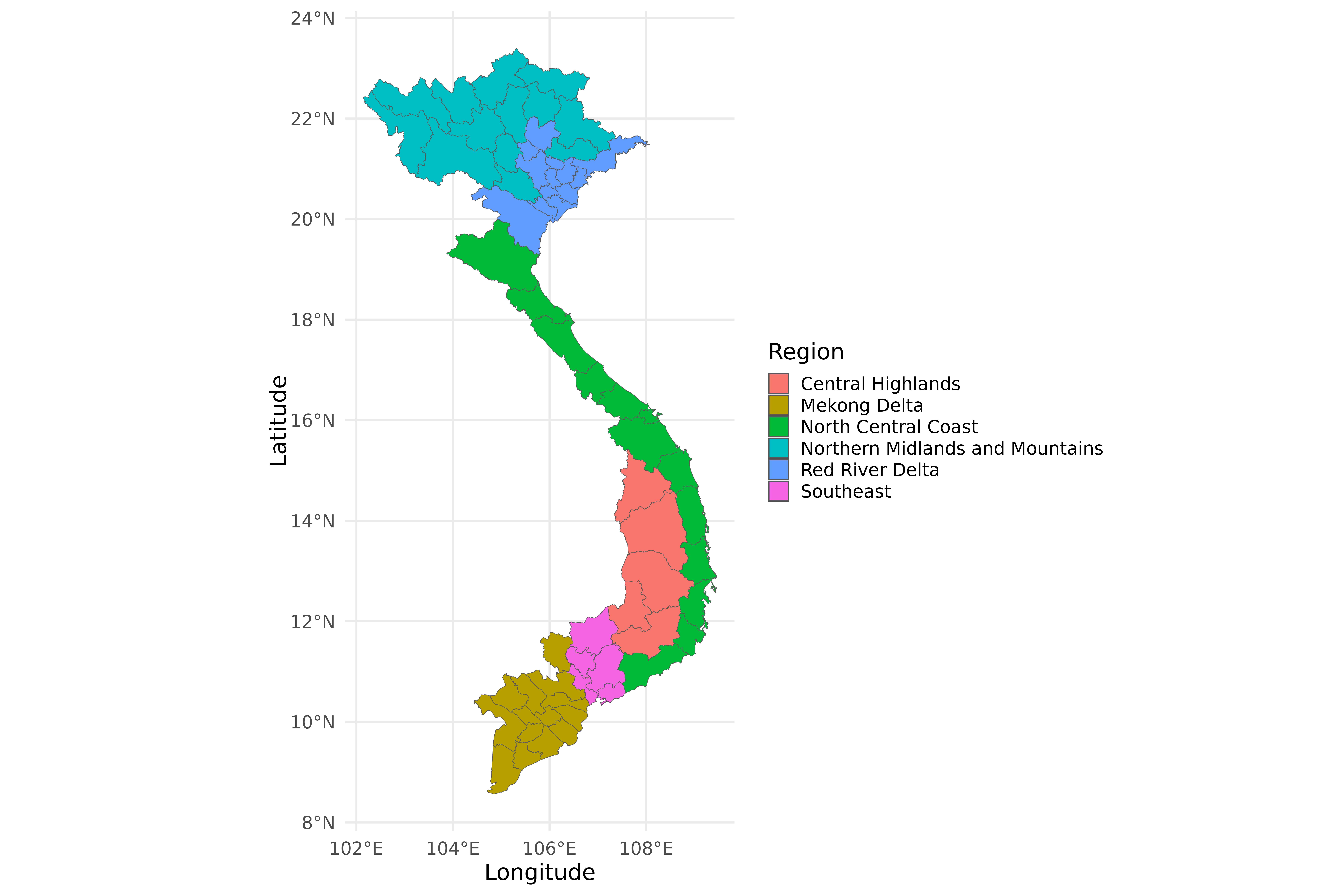
Map of Vietnam by region.
vntr <- vnt |>
select(!province) |>
group_by(region_name) |>
summarise(t_max = mean(t_max))
vntr |> plot_funs(t_max, color = region_name) +
geom_hline(yintercept = 1 / diff(vnt$t_max[[1]]$basis$rangeval), linetype = "dotted") +
geom_function(fun = function(x) eval(mean(c(vnt$t_max)), x), color = "black", linetype = "dotted", linewidth = 1) +
theme_legend_inside +
labs(
x = "Temperature (deg. Celsius)",
y = "Density",
color = "Region"
)
#> Warning: Multiple drawing groups in `geom_function()`
#> i Did you use the correct group, colour, or fill aesthetics?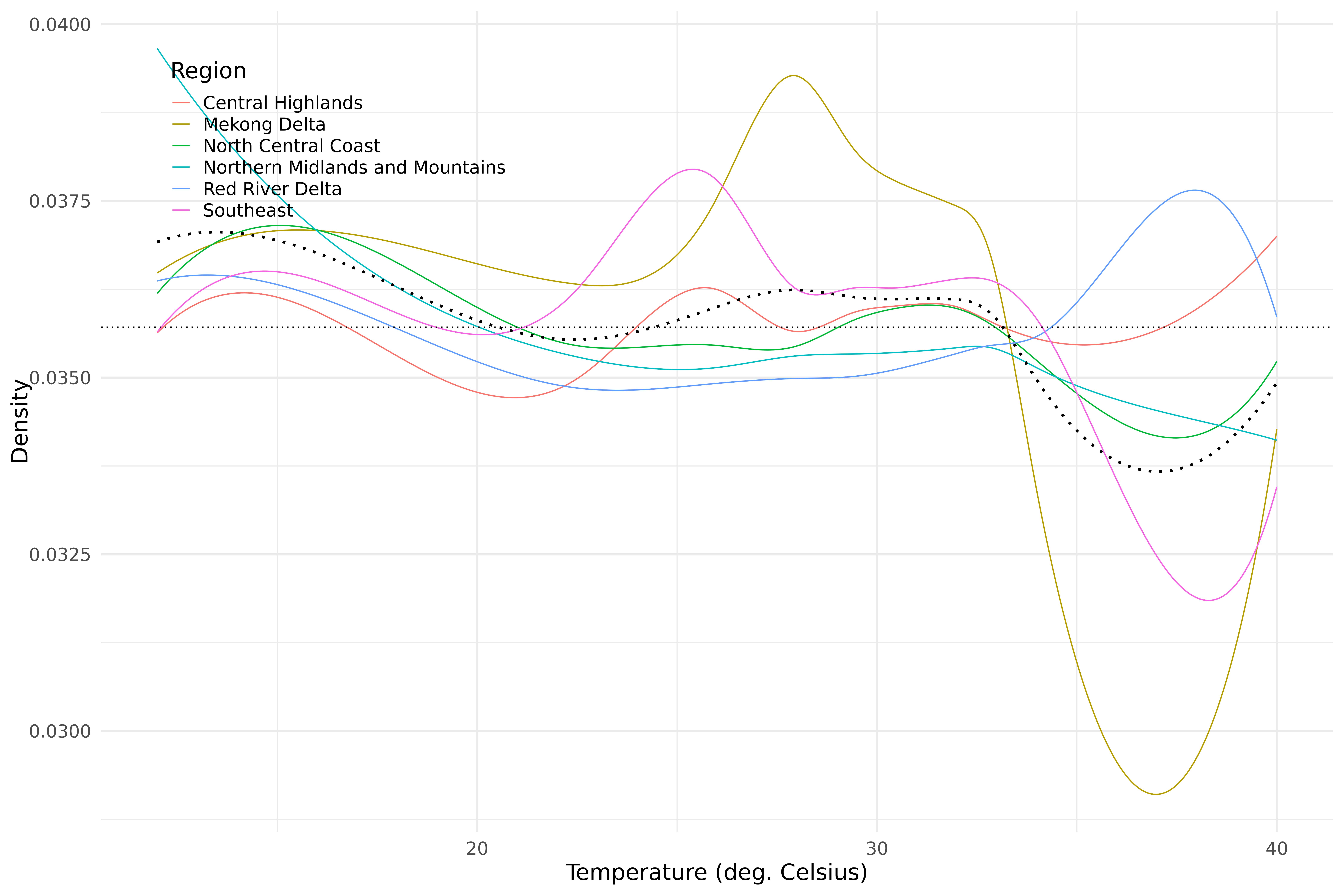
Mean trend density of maximum temperature per region.
vntry <- vietnam_temperature_dd |>
left_join(vietnam_regions, by = c("region" = "code")) |>
rename(region_name = name) |>
arrange(region, province) |>
select(!province) |>
group_by(region_name, year) |>
summarise(t_max = mean(t_max))
#> `summarise()` has grouped output by 'region_name'. You can override using the
#> `.groups` argument.
vntry |> plot_funs(t_max, color = region_name, linewidth = year, alpha = year) +
facet_grid(vars(str_wrap(region_name, 14)), scales = "free_y") +
scale_linewidth(range = c(0.2, 2)) +
theme_legend_inside +
labs(
x = "Temperature (deg. Celsius)",
y = "Density",
color = "Region"
)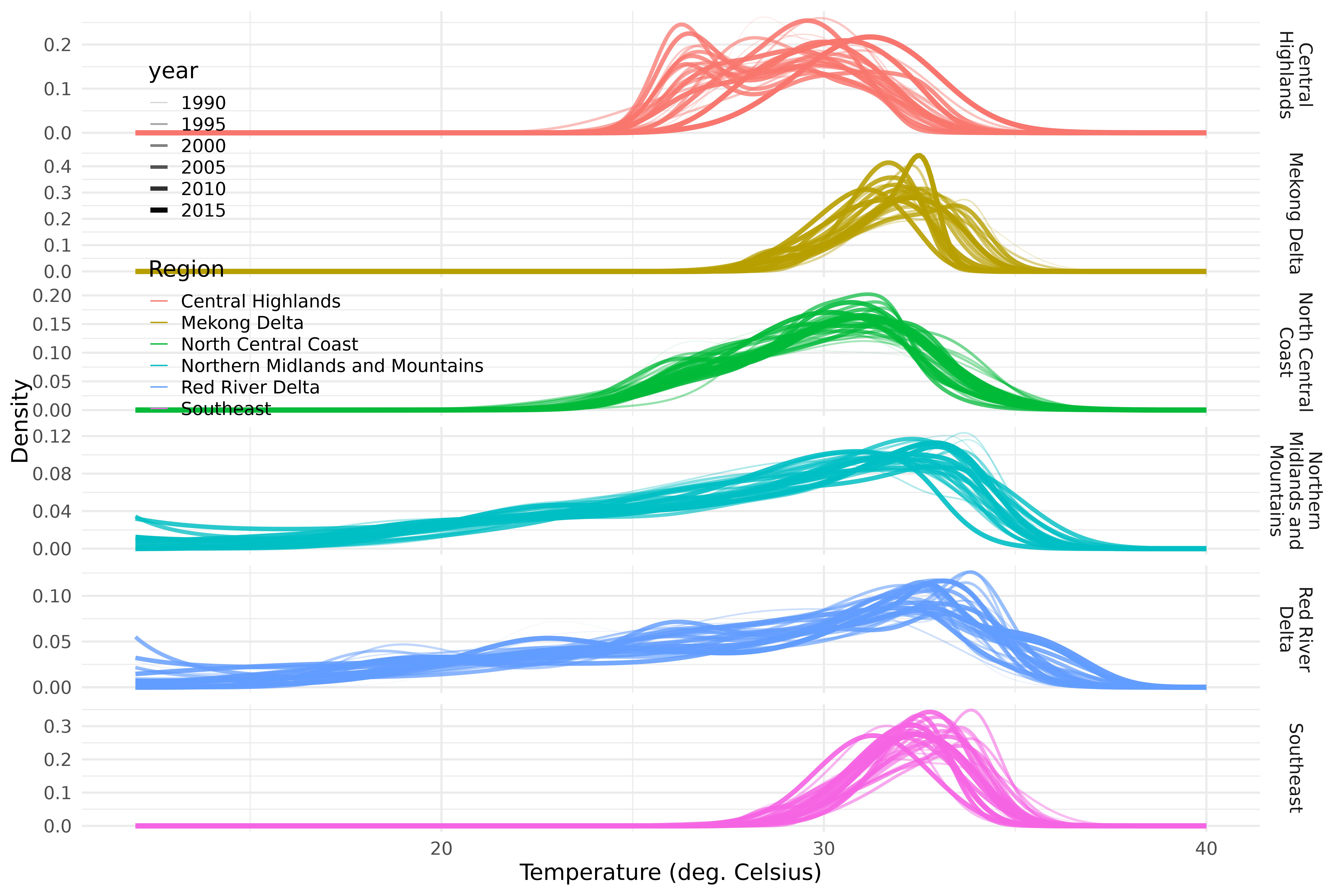
Mean trend density of maximum temperature per region.
Preliminary study
Normality assumption
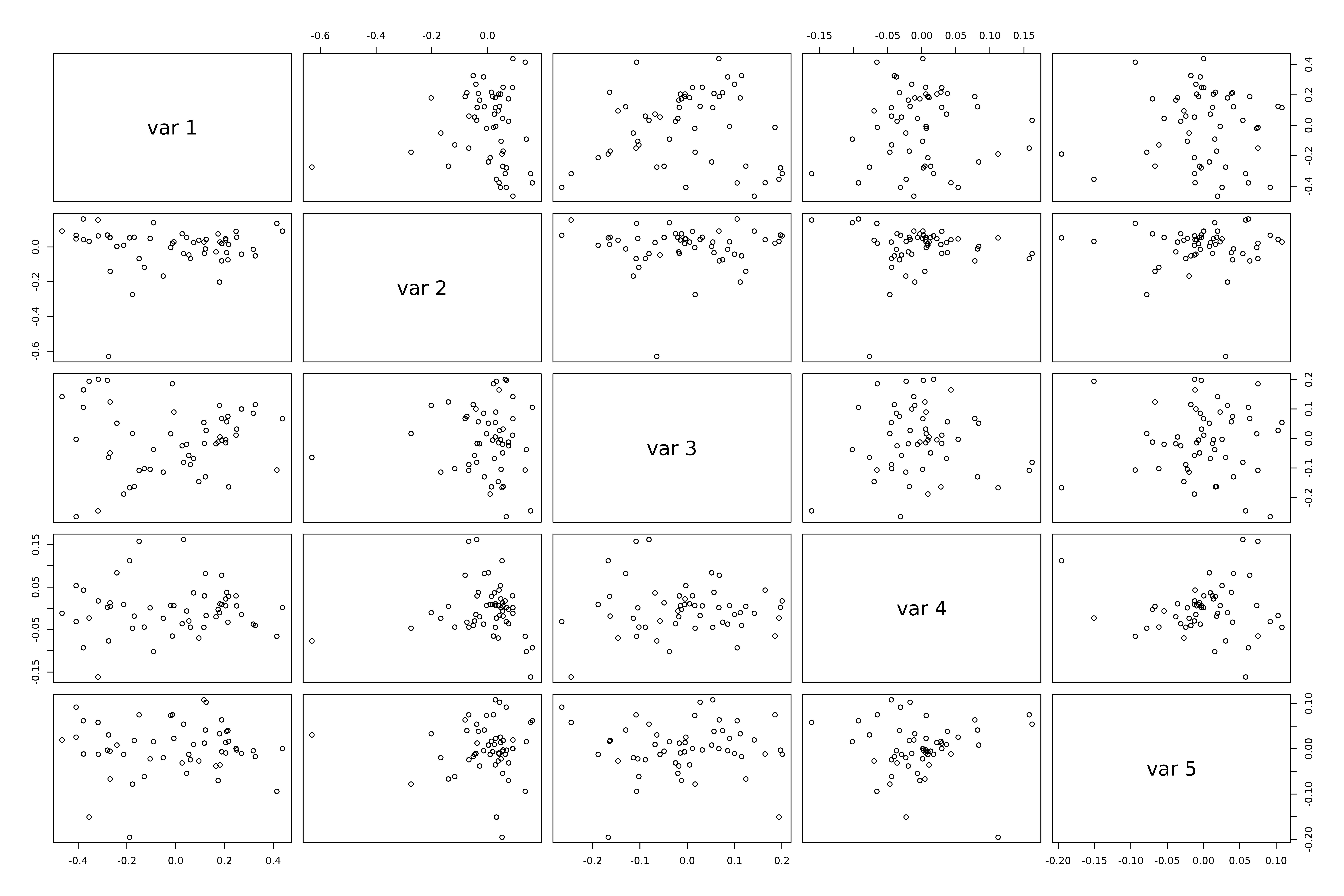
mvn(vnt_pc$scores, mvnTest = "mardia")
#> $multivariateNormality
#> Test Statistic p value Result
#> 1 Mardia Skewness 236.796233340498 8.36360275201567e-32 NO
#> 2 Mardia Kurtosis 11.0447074304137 0 NO
#> 3 MVN <NA> <NA> NO
#>
#> $univariateNormality
#> Test Variable Statistic p value Normality
#> 1 Anderson-Darling Column1 1.4247 0.001 NO
#> 2 Anderson-Darling Column2 3.5984 <0.001 NO
#> 3 Anderson-Darling Column3 0.2838 0.6209 YES
#> 4 Anderson-Darling Column4 1.1746 0.0042 NO
#> 5 Anderson-Darling Column5 1.3594 0.0015 NO
#>
#> $Descriptives
#> n Mean Std.Dev Median Min Max 25th
#> 1 63 -3.271752e-18 0.23634837 0.054066924 -0.4658859 0.4379686 -0.22698516
#> 2 63 -3.940975e-18 0.11303792 0.021633456 -0.6304448 0.1611768 -0.03712273
#> 3 63 2.979835e-18 0.10885494 -0.005430535 -0.2651669 0.2007059 -0.06621715
#> 4 63 4.553004e-19 0.05495679 0.002209999 -0.1616571 0.1619070 -0.03196695
#> 5 63 -2.204746e-18 0.05252636 -0.003134425 -0.1954665 0.1080602 -0.01736823
#> 75th Skew Kurtosis
#> 1 0.18922297 -0.2551881 -1.1623638
#> 2 0.05474903 -2.9836297 13.4528218
#> 3 0.07126844 -0.1405793 -0.3803929
#> 4 0.01976211 0.3854705 1.6145156
#> 5 0.02425947 -0.8676932 2.5030284Homogeneity of variances
vnt_var <- var.fd(c(as.fd(vnt$t_max)))
tt <- seq(
vnt_var$sbasis$rangeval[1],
vnt_var$sbasis$rangeval[2],
length.out = 401
)
z <- eval.bifd(tt, tt, vnt_var) # Covariance
z <- cor.fd(tt, c(as.fd(vnt$t_max))) # Correlation
df <- data.frame(
x = rep(tt, each = length(tt)),
y = rep(tt, times = length(tt)),
z = as.vector(z)
)
ggplot(df, aes(x = x, y = y, z = z)) +
geom_contour_filled() +
labs(
fill = "Correlation",
x = "Temperature (deg. Celsius)", y = "Temperature (deg. Celsius)"
)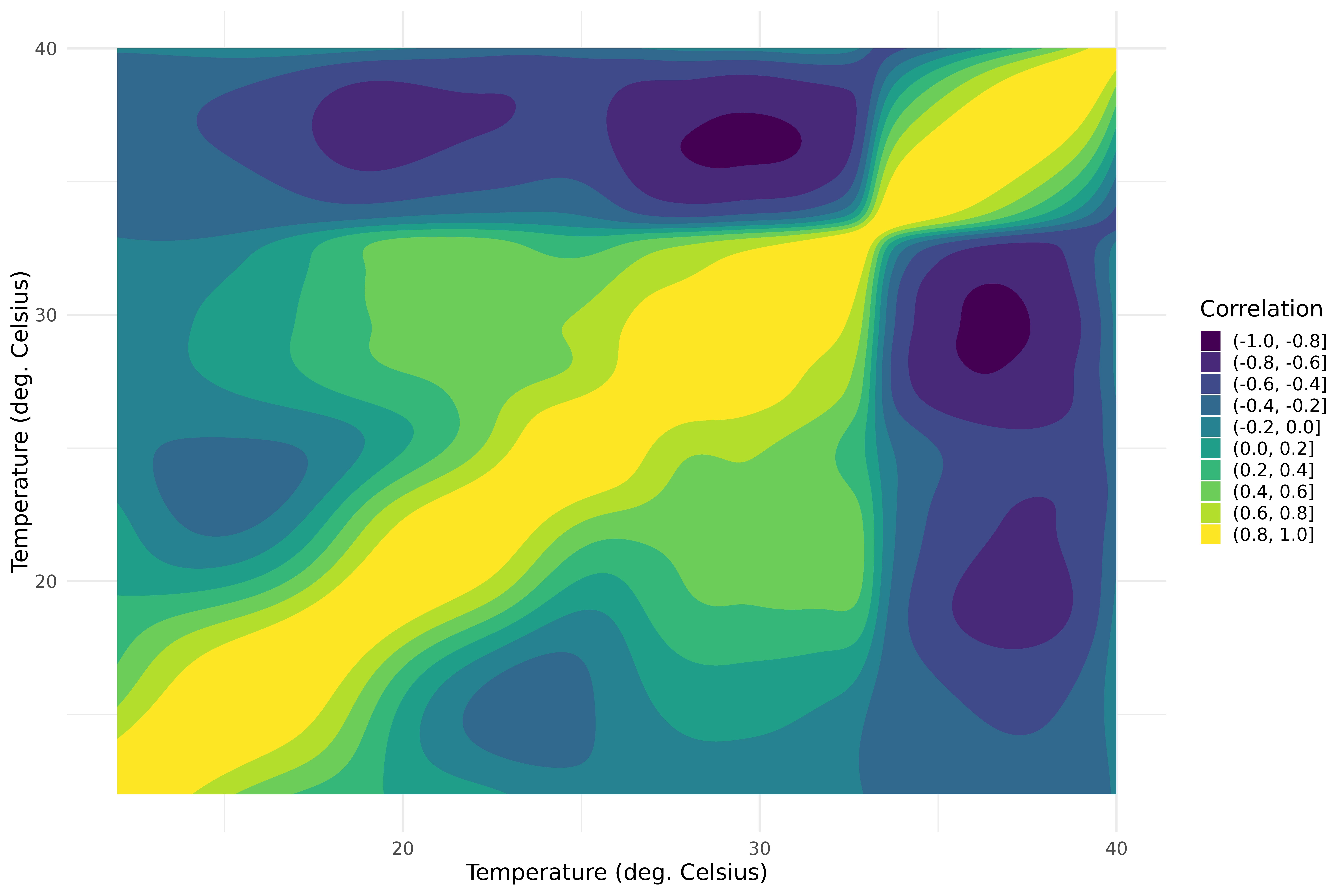
Correlation function within the MDR region.
Global tests
Distributional ANOVA
vnte <- vnt |>
mutate(t_max = as.fd(t_max)) |>
eval_funs(t_max, n = 1401) |>
select(province, x, y) |>
pivot_wider(names_from = province, values_from = y) |>
column_to_rownames("x")
vnt_province_region <- vietnam_provinces$region
names(vnt_province_region) <- vietnam_provinces$province
plotFANOVA(vnte, vnt_province_region[colnames(vnte)], separately = TRUE)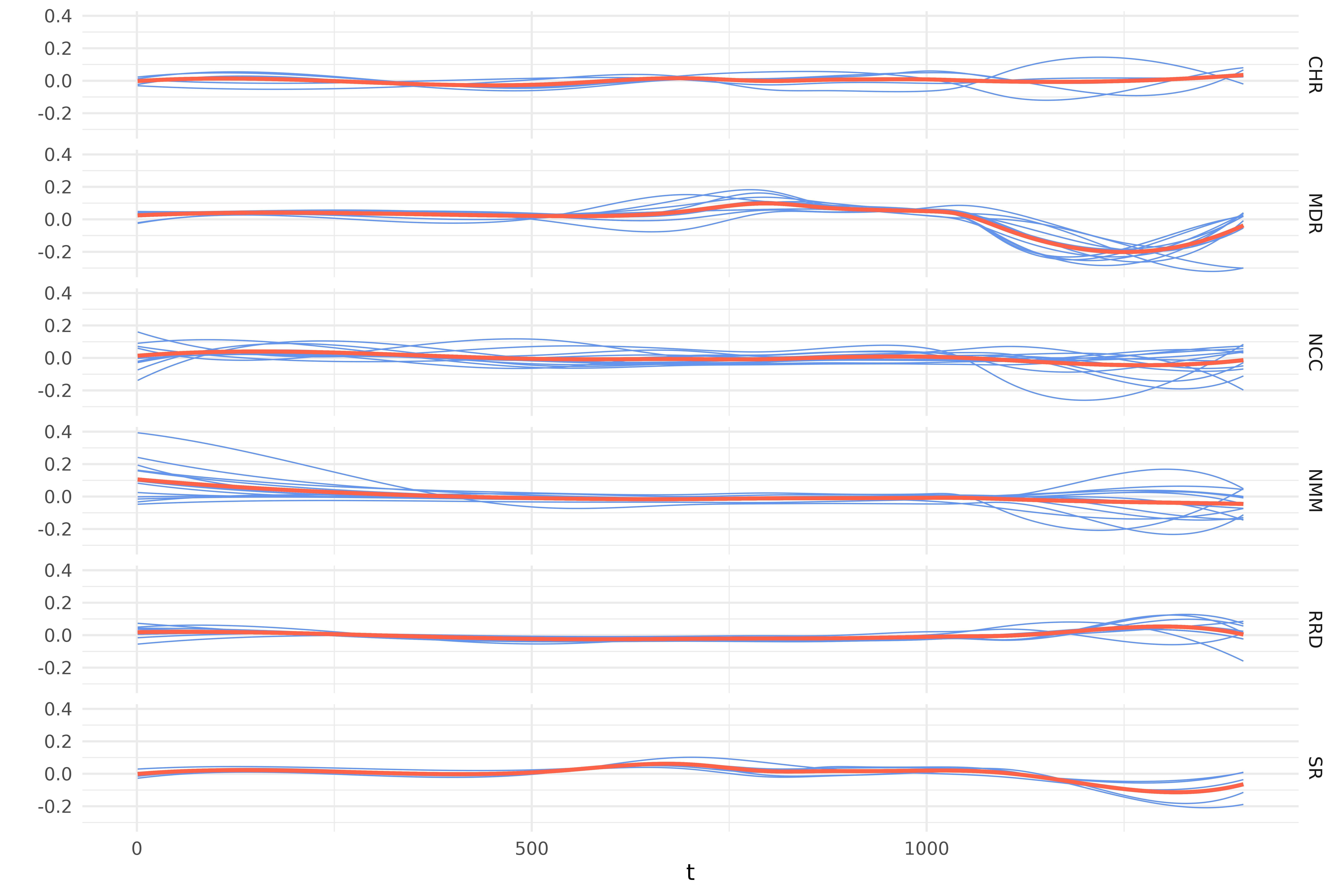
stat_list <- list("GPF", "Fmaxb", "CS", "L2B", "FB")
res <- fanova.tests(vnte, vnt_province_region[colnames(vnte)],
test = stat_list,
params = list(
paramCS = 100,
paramFmaxb = 100,
paramTRP = list(B.TRP = 100)
)
)
res_tbl <- res[as_vector(stat_list)] |>
sapply(function(x) c(`Statistic` = x$stat, `p-value` = x$pval)) |>
t() |>
as.data.frame() |>
rownames_to_column("Name") |>
kable_format()
res_tbl
#> Name Statistic p-value
#> 1 GPF 10 0e+00 ***
#> 2 Fmaxb 36 0e+00 ***
#> 3 CS 740 0e+00 ***
#> 4 L2B 130 0e+00 ***
#> 5 FB 9.9 0e+00 ***#> ```tex
#> \begin{table}
#> \centering
#> \begin{tabular}[t]{lll}
#> \toprule
#> Name & Statistic & p-value\\
#> \midrule
#> \cellcolor{gray!10}{GPF} & \cellcolor{gray!10}{10} & \cellcolor{gray!10}{0e+00 ***}\\
#> Fmaxb & 36 & 0e+00 ***\\
#> \cellcolor{gray!10}{CS} & \cellcolor{gray!10}{740} & \cellcolor{gray!10}{0e+00 ***}\\
#> L2B & 130 & 0e+00 ***\\
#> \cellcolor{gray!10}{FB} & \cellcolor{gray!10}{9.9} & \cellcolor{gray!10}{0e+00 ***}\\
#> \bottomrule
#> \end{tabular}
#> \end{table}
#> ```Pairwise comparisons
vntp <- t(combn(c("NMM", "RRD", "NCC", "CHR", "SR", "MDR"), m = 2))
colnames(vntp) <- c("region1", "region2")
pairwise_anova <- function(region1, region2) {
data <- vnte[vnt_province_region[colnames(vnte)] %in% c(region1, region2)]
res <- fanova.tests(data, vnt_province_region[colnames(data)],
test = stat_list,
params = list(
paramCS = 100,
paramFmaxb = 100,
paramTRP = list(B.TRP = 100)
)
)
res[as_vector(stat_list)] |>
sapply(function(x) c(x$pval)) |> c()
}
res_tbl <- as_tibble(vntp) |>
rowwise() |>
mutate(res = map2(region1, region2, pairwise_anova)) |>
unnest_wider(res) |>
kable_format()
res_tbl
#> # A tibble: 15 x 7
#> region1 region2 GPF Fmaxb CS L2B FB
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 NMM RRD "2.6e-02 **" "0.12 " "0e+00 ***" "7e-03 ***" "1.3e-02~
#> 2 NMM NCC "0.53 " "0.33 " "0.45 " "0.44 " "0.46 "
#> 3 NMM CHR "4.2e-02 **" "2e-02 **" "0.17 " "0.21 " "0.25 "
#> 4 NMM SR "2e-06 ***" "0e+00 ***" "2e-02 **" "8e-02 *" "0.11 "
#> 5 NMM MDR "0e+00 ***" "0e+00 ***" "0e+00 ***" "3.6e-10 ***" "9.2e-08~
#> 6 RRD NCC "1.9e-03 ***" "1e-02 ***" "0e+00 ***" "1.7e-04 ***" "5.8e-04~
#> 7 RRD CHR "2.2e-03 ***" "0e+00 ***" "0.23 " "0.11 " "0.14 "
#> 8 RRD SR "0e+00 ***" "0e+00 ***" "0e+00 ***" "5.8e-12 ***" "2.6e-07~
#> 9 RRD MDR "0e+00 ***" "0e+00 ***" "0e+00 ***" "0e+00 ***" "0e+00 *~
#> 10 NCC CHR "0.24 " "0.17 " "0.24 " "0.29 " "0.32 "
#> 11 NCC SR "4.3e-02 **" "1e-02 ***" "1e-02 ***" "5.9e-02 *" "8e-02 *"
#> 12 NCC MDR "1.3e-14 ***" "0e+00 ***" "0e+00 ***" "5.3e-14 ***" "1.8e-10~
#> 13 CHR SR "2.1e-02 **" "5e-02 **" "4e-02 **" "1.1e-02 **" "3.4e-02~
#> 14 CHR MDR "3.2e-12 ***" "0e+00 ***" "0e+00 ***" "2.4e-12 ***" "1.5e-08~
#> 15 SR MDR "1.8e-07 ***" "0e+00 ***" "0e+00 ***" "6e-05 ***" "4.1e-04~#> ```tex
#> \begin{table}
#> \centering
#> \begin{tabular}[t]{lllllll}
#> \toprule
#> region1 & region2 & GPF & Fmaxb & CS & L2B & FB\\
#> \midrule
#> \cellcolor{gray!10}{NMM} & \cellcolor{gray!10}{RRD} & \cellcolor{gray!10}{2.6e-02 **} & \cellcolor{gray!10}{0.12} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{7e-03 ***} & \cellcolor{gray!10}{1.3e-02 **}\\
#> NMM & NCC & 0.53 & 0.33 & 0.45 & 0.44 & 0.46\\
#> \cellcolor{gray!10}{NMM} & \cellcolor{gray!10}{CHR} & \cellcolor{gray!10}{4.2e-02 **} & \cellcolor{gray!10}{2e-02 **} & \cellcolor{gray!10}{0.17} & \cellcolor{gray!10}{0.21} & \cellcolor{gray!10}{0.25}\\
#> NMM & SR & 2e-06 *** & 0e+00 *** & 2e-02 ** & 8e-02 * & 0.11\\
#> \cellcolor{gray!10}{NMM} & \cellcolor{gray!10}{MDR} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{3.6e-10 ***} & \cellcolor{gray!10}{9.2e-08 ***}\\
#> \addlinespace
#> RRD & NCC & 1.9e-03 *** & 1e-02 *** & 0e+00 *** & 1.7e-04 *** & 5.8e-04 ***\\
#> \cellcolor{gray!10}{RRD} & \cellcolor{gray!10}{CHR} & \cellcolor{gray!10}{2.2e-03 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0.23} & \cellcolor{gray!10}{0.11} & \cellcolor{gray!10}{0.14}\\
#> RRD & SR & 0e+00 *** & 0e+00 *** & 0e+00 *** & 5.8e-12 *** & 2.6e-07 ***\\
#> \cellcolor{gray!10}{RRD} & \cellcolor{gray!10}{MDR} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0e+00 ***}\\
#> NCC & CHR & 0.24 & 0.17 & 0.24 & 0.29 & 0.32\\
#> \addlinespace
#> \cellcolor{gray!10}{NCC} & \cellcolor{gray!10}{SR} & \cellcolor{gray!10}{4.3e-02 **} & \cellcolor{gray!10}{1e-02 ***} & \cellcolor{gray!10}{1e-02 ***} & \cellcolor{gray!10}{5.9e-02 *} & \cellcolor{gray!10}{8e-02 *}\\
#> NCC & MDR & 1.3e-14 *** & 0e+00 *** & 0e+00 *** & 5.3e-14 *** & 1.8e-10 ***\\
#> \cellcolor{gray!10}{CHR} & \cellcolor{gray!10}{SR} & \cellcolor{gray!10}{2.1e-02 **} & \cellcolor{gray!10}{5e-02 **} & \cellcolor{gray!10}{4e-02 **} & \cellcolor{gray!10}{1.1e-02 **} & \cellcolor{gray!10}{3.4e-02 **}\\
#> CHR & MDR & 3.2e-12 *** & 0e+00 *** & 0e+00 *** & 2.4e-12 *** & 1.5e-08 ***\\
#> \cellcolor{gray!10}{SR} & \cellcolor{gray!10}{MDR} & \cellcolor{gray!10}{1.8e-07 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{0e+00 ***} & \cellcolor{gray!10}{6e-05 ***} & \cellcolor{gray!10}{4.1e-04 ***}\\
#> \bottomrule
#> \end{tabular}
#> \end{table}
#> ```